As this RNA‑first biotech scaled its discovery programs, the science moved faster than the data. SPR, mass spectrometry, and registry information lived in different tools and formats, which made it hard to trace lineage, keep teams aligned, and answer simple questions with confidence. Scientists spent too much time piecing together exports and too little time reviewing results. The lack of SPR integration and mass spec integration meant critical context was frequently trapped in silos rather than flowing through a common model. What the company needed wasn’t another monolithic system—it needed a connective layer that would unify how data flows, how it’s described, and how people interact with it.
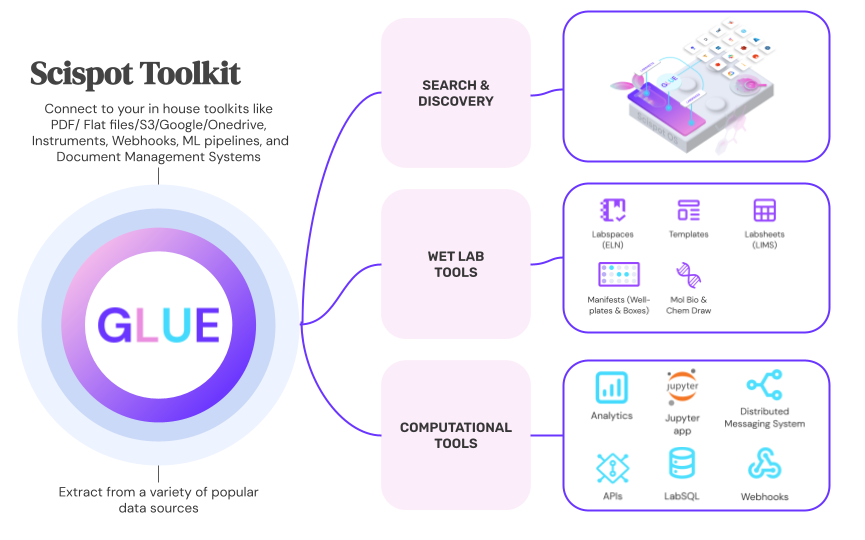
Scispot GLUE became that connective tissue. The company established a single, canonical way to represent its biology—targets, sub‑targets, DNA templates, IVT artifacts, RNA constructs, and allowed modifications—so everyone referenced the same entities the same way. With that shared language in place, GLUE orchestrated the movement of data across SPR and mass spec: runs came in, were standardized against the model, presented for review in a consistent workspace, and then packaged for downstream systems with dependable structure and context. In practice, GLUE served as SPR data processing software while also enabling mass spectrometry data automation, tying instruments and analyses back to the same identifiers. The “how” is intentionally simple: self‑service inputs, event‑driven transformations, human‑in‑the‑loop review, and predictable outputs. The result is not a new workflow for scientists to learn, but a reliable backbone behind the workflows they already use.

The impact showed up quickly. Biophysics and analytical teams began working to the same cadence and conventions, which reduced hand‑offs and rework. Registry updates no longer introduced drift because constructs and modifications were chosen from controlled terms rather than typed free‑form. Historical data was brought into the same model, so new experiments could be interpreted alongside old ones without fragile spreadsheets or guesswork. When reviews surfaced an anomaly, it was fixed once in the pipeline and stayed fixed everywhere thereafter. Leaders gained traceability from report back to sample, and reviewers saw the same context—curves, parameters, notes—without chasing files. The registry layer doubled as RNA construct registry software, ensuring constructs, modifications, and lineage remained synchronized with both SPR integration and mass spec integration.
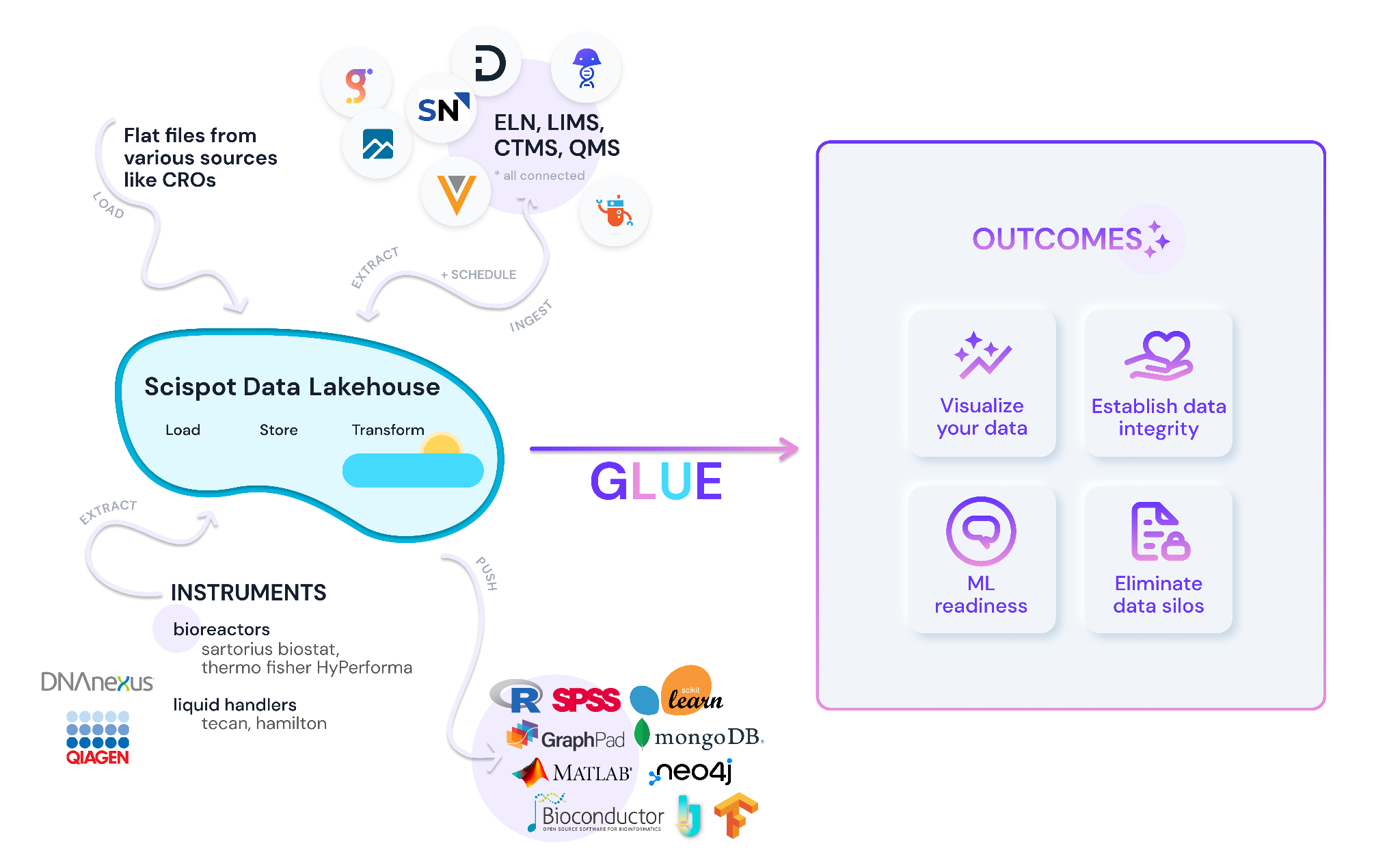
Compliance and readiness improved as a by‑product of this design. Every run carries provenance, every transformation is recorded, and every published artifact can be regenerated consistently. Role‑appropriate permissions and single sign‑on protect sensitive work without slowing it down. Most importantly, the organization now produces clean, linked, analysis‑ready data by default, which accelerates routine decision‑making and lays a foundation for cross‑assay analytics and AI. Because GLUE centralizes transformations and approvals, updates to processing logic for SPR or mass spectrometry propagate system‑wide, reinforcing both SPR data processing software best practices and scalable mass spectrometry data automation.

The lessons are broadly applicable: agree on the schema before you automate; make scientific review a first‑class step rather than an afterthought; standardize names and packaging early; support late edits without forcing full reprocessing; and treat versioning and lineage as system properties, not heroic efforts. From here, the company is extending the same pattern to additional instruments, promoting specialized calculations into validated platform functions, and deepening API handoffs to its discovery data platforms. The science keeps moving—and now the data infrastructure moves with it. If you’re evaluating next steps, explore how Scispot enables SPR integration and mass spec integration to unify instruments, registries, and analyses into a single, interoperable layer.





.webp)
.webp)
.webp)



